P Systems Implementation: A Model of Computing for Biological Mitochondrial Rules using Object Oriented Programming
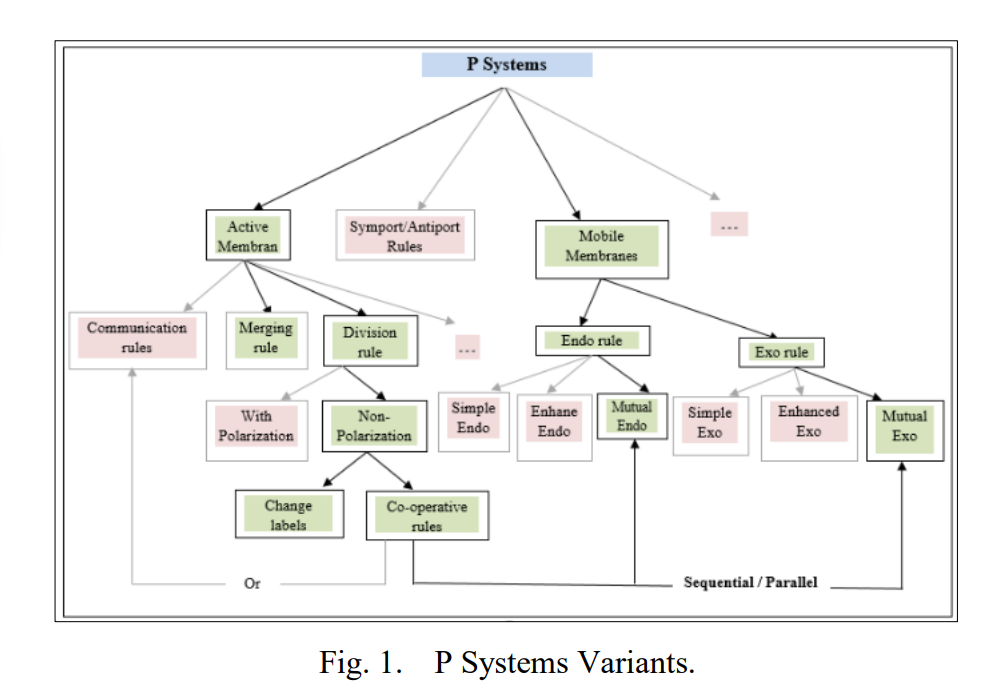
Membrane computing is a computational framework that depends on the behavior and structure of living cells. P systems are arising from the biological processes which occur in the living cells’ organelles in a non-deterministic and maximally parallel manner. This paper aims to build a powerful computational model that combines the rules of active and mobile membranes, called Mutual Dynamic Membranes (MDM). The proposed model will describe the biological mechanisms of the metabolic regulation of mitochondrial dynamics made by mitochondrial membranes. The behaviors of the proposed model regulate the mitochondrial fusion and fission processes based on the combination of P systems variants. The combination of different variants in our computational model and their high parallelism lead to provide the possibility for solving problems that belong to NP-complete classes in polynomial time in a more efficient way than other conventional methods. To evaluate this model, it was applied to solve the SAT problem and calculate a set of computational complexity results that approved the quality of our model. Another contribution of this paper, the biological models of mitochondrial is presented in the formal class relationship diagrams were designed and illustrated using Unified Modeling Language (UML). This mechanism will be used to define a new specification of membrane processes into Object-Oriented Programming (OOP) to add the functionality of a common programming methodology to solve a large category of NP-hard problems as an interesting initiative of future research. © 2021