Parallelizing exact motif finding algorithms on multi-core
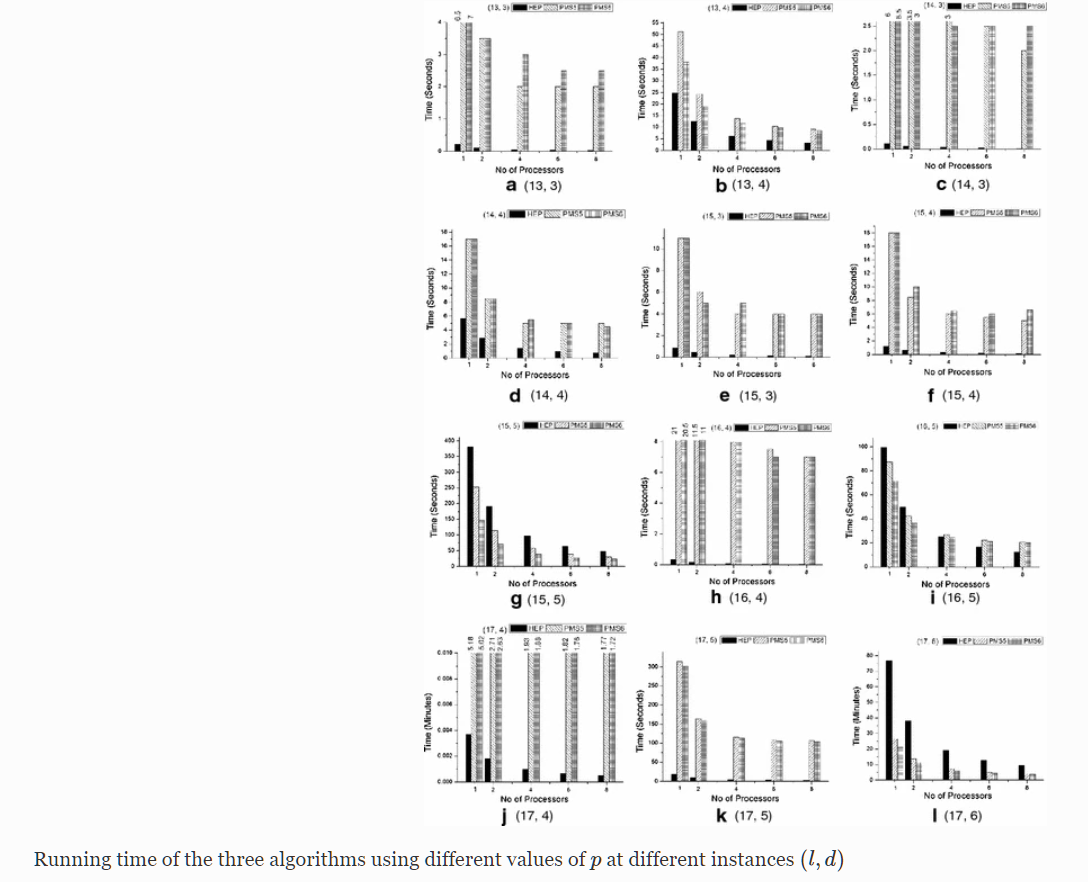
The motif finding problem is one of the important and challenging problems in bioinformatics. A variety of sequential algorithms have been proposed to find exact motifs, but the running time is still not suitable due to high computational complexity of finding motifs. In this paper we parallelize three efficient sequential algorithms which are HEPPMSprune, PMS5 and PMS6. We implement the algorithms on a Dual Quad-Core machine using openMP to measure the performance of each algorithm. Our experiment on simulated data show that: (1) the parallel PMS6 is faster than the other algorithms in case of challenging instances, while the parallel HEPPMSprune is faster than the other algorithms in most of solvable instances; (2) the scalability of parallel HEPPMSprune is linear for all instances, while the scalability of parallel PMS5 and PMS6 is linear in case of challenging instances only; (3) the memory used by HEPPMSprune is less than that of the other algorithms. © 2014, Springer Science+Business Media New York.